AuroraGPT: Foundation Models for Science
@ Foundation Models for the Electric Grid
Sam Foreman
2025-02-12
🎯 AuroraGPT: Goals
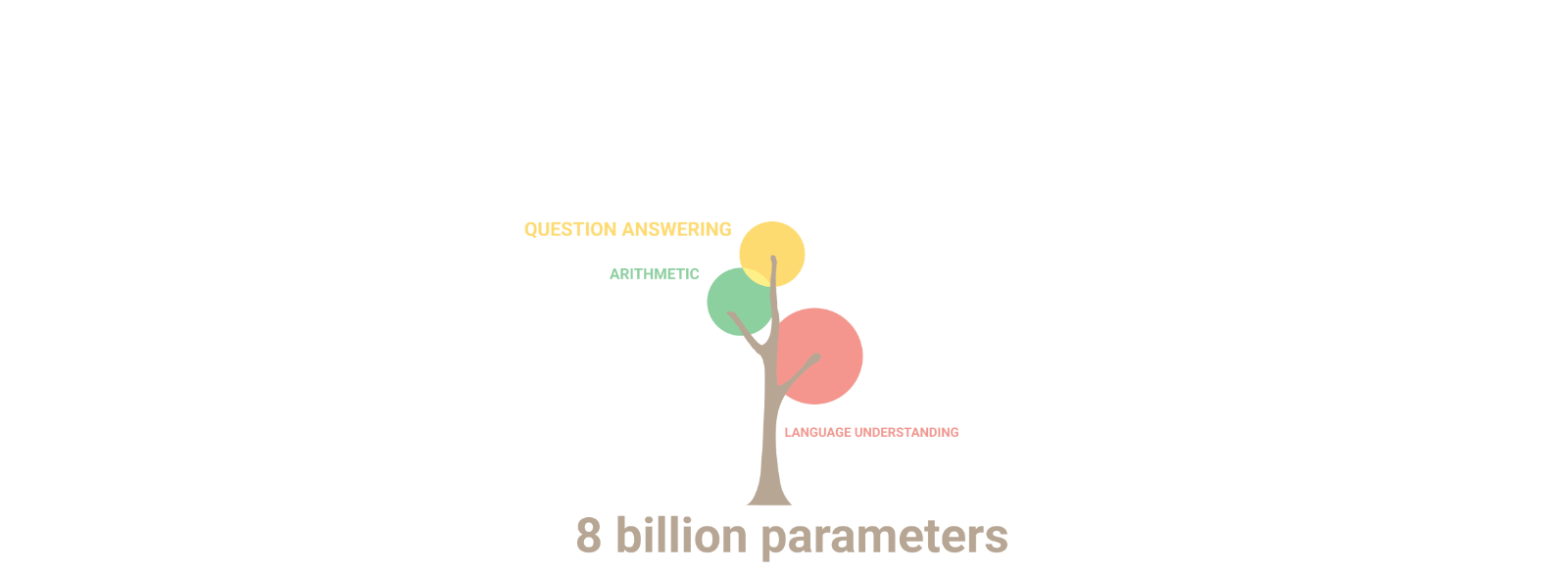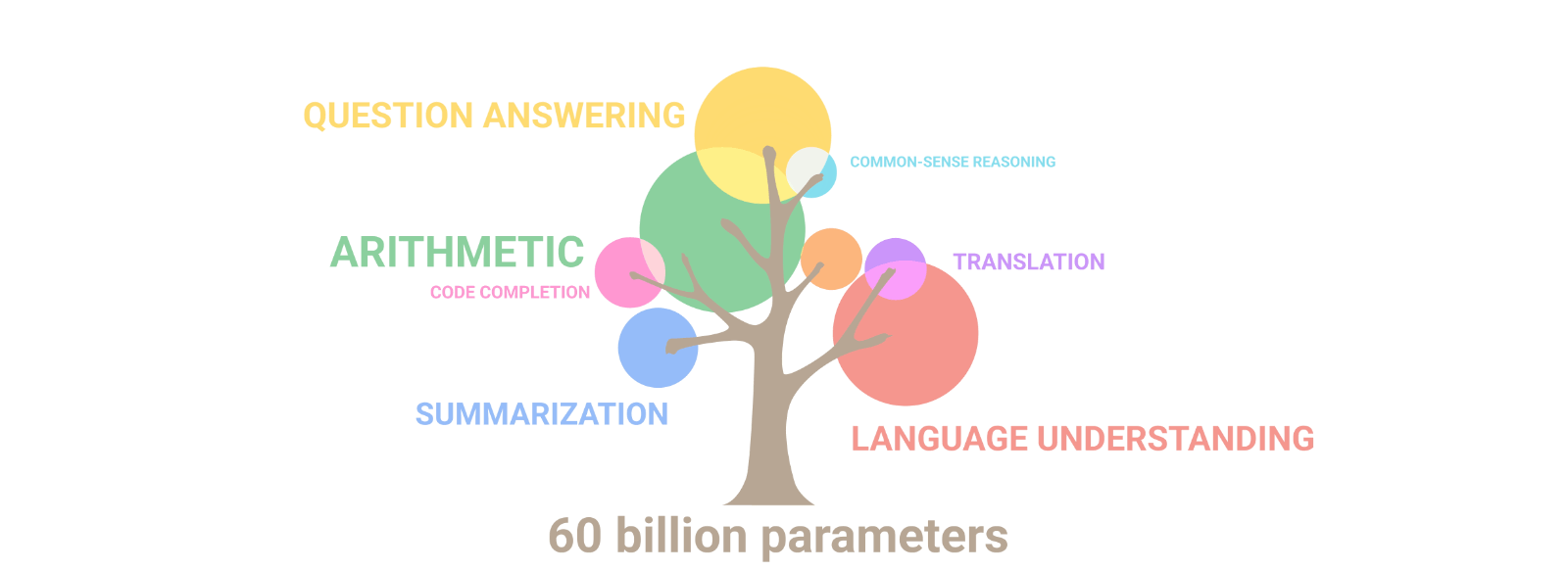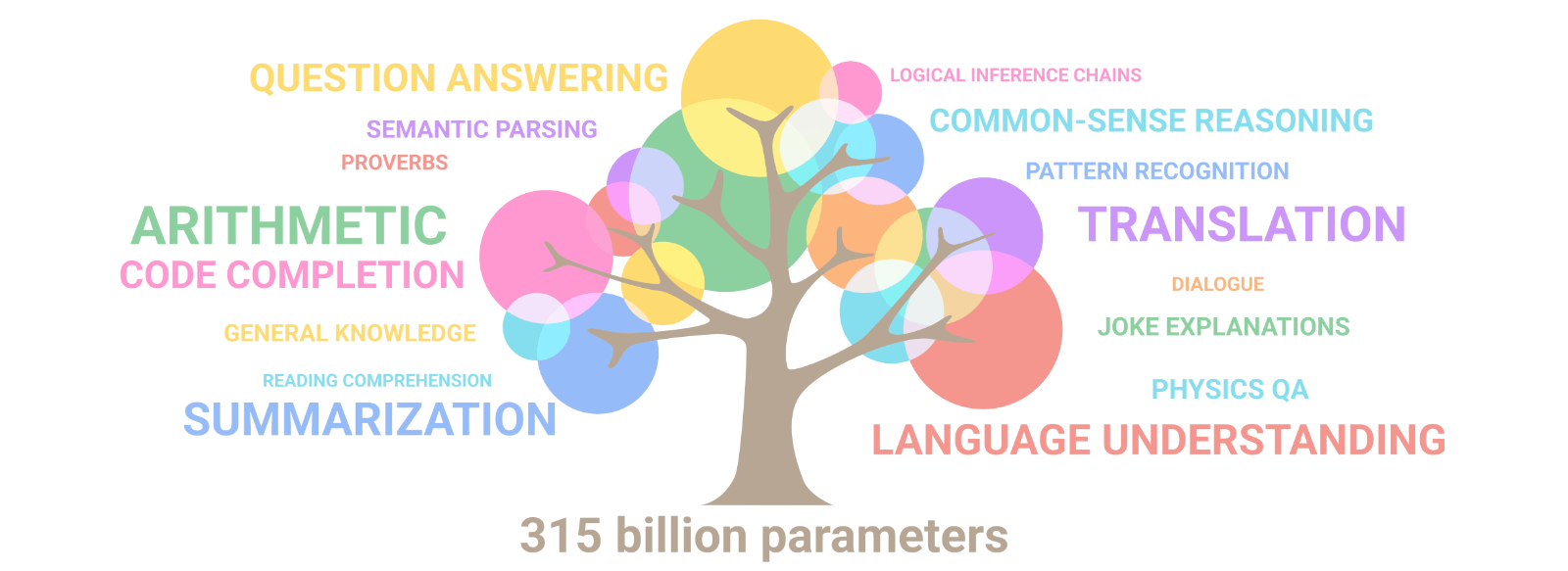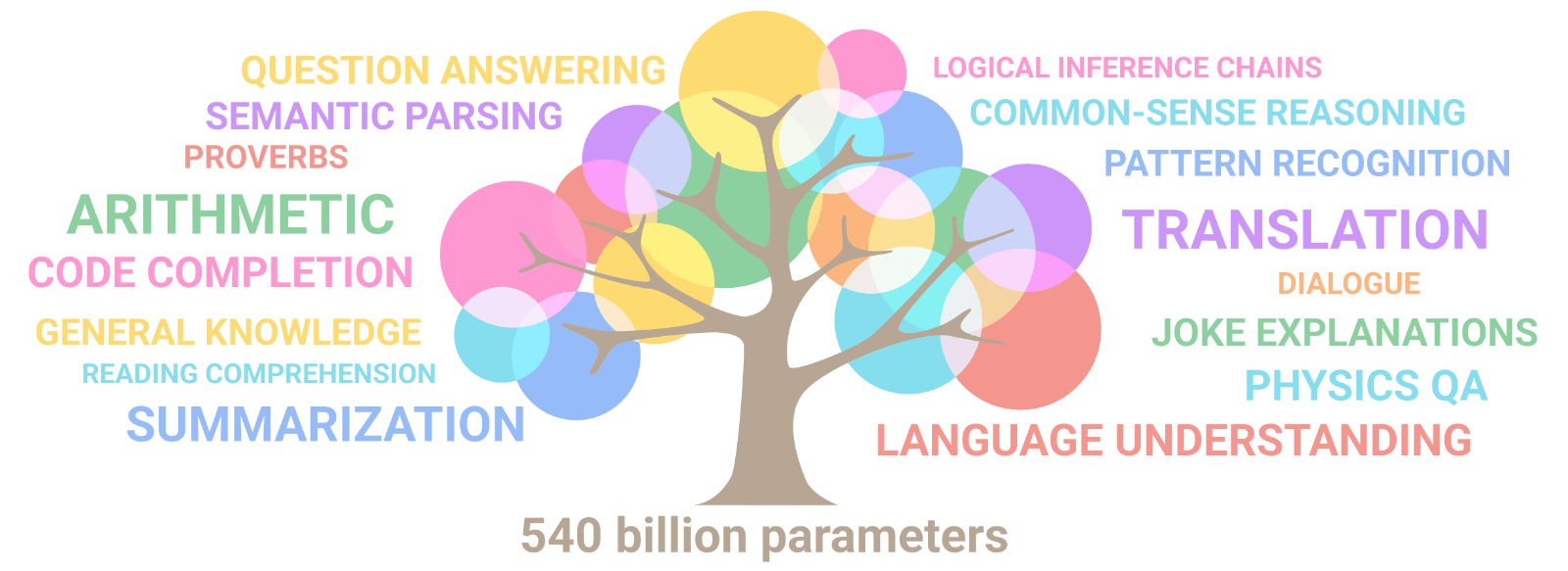
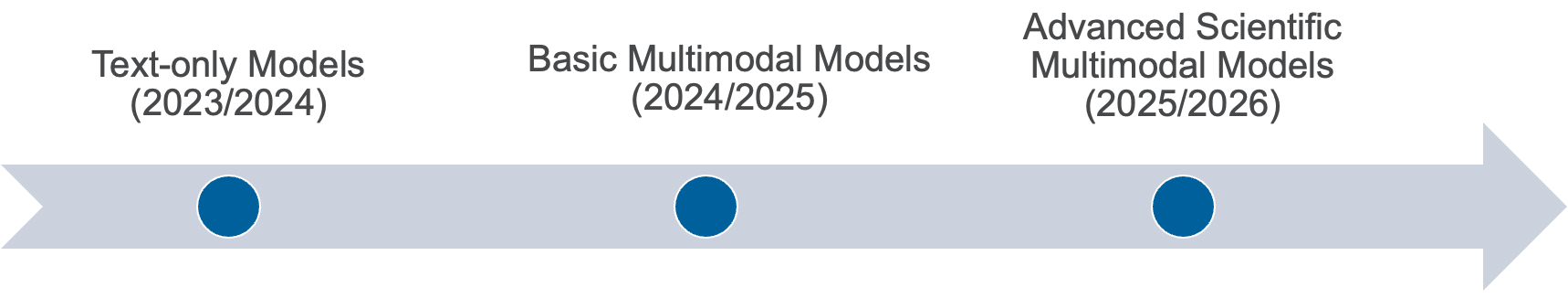
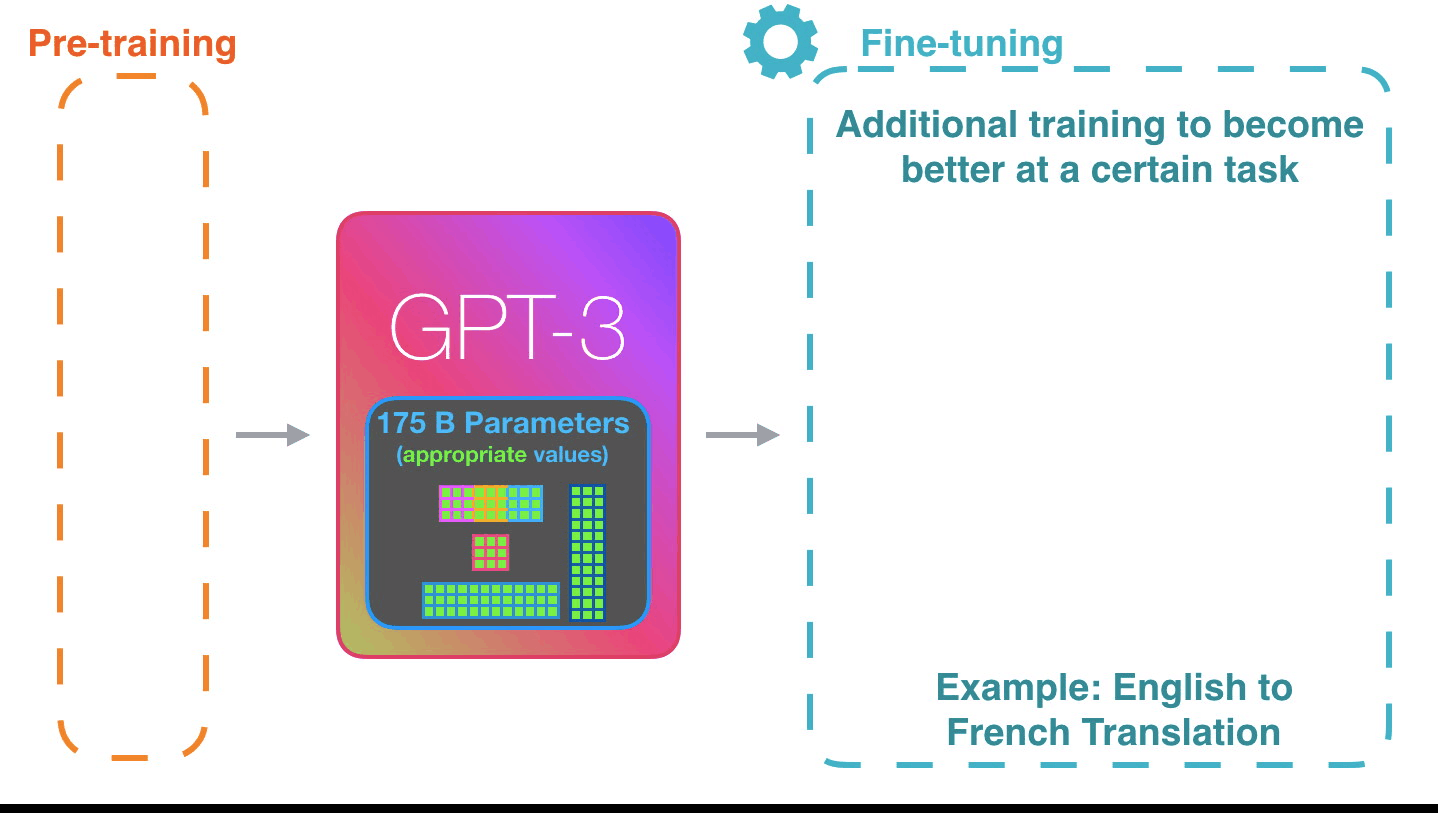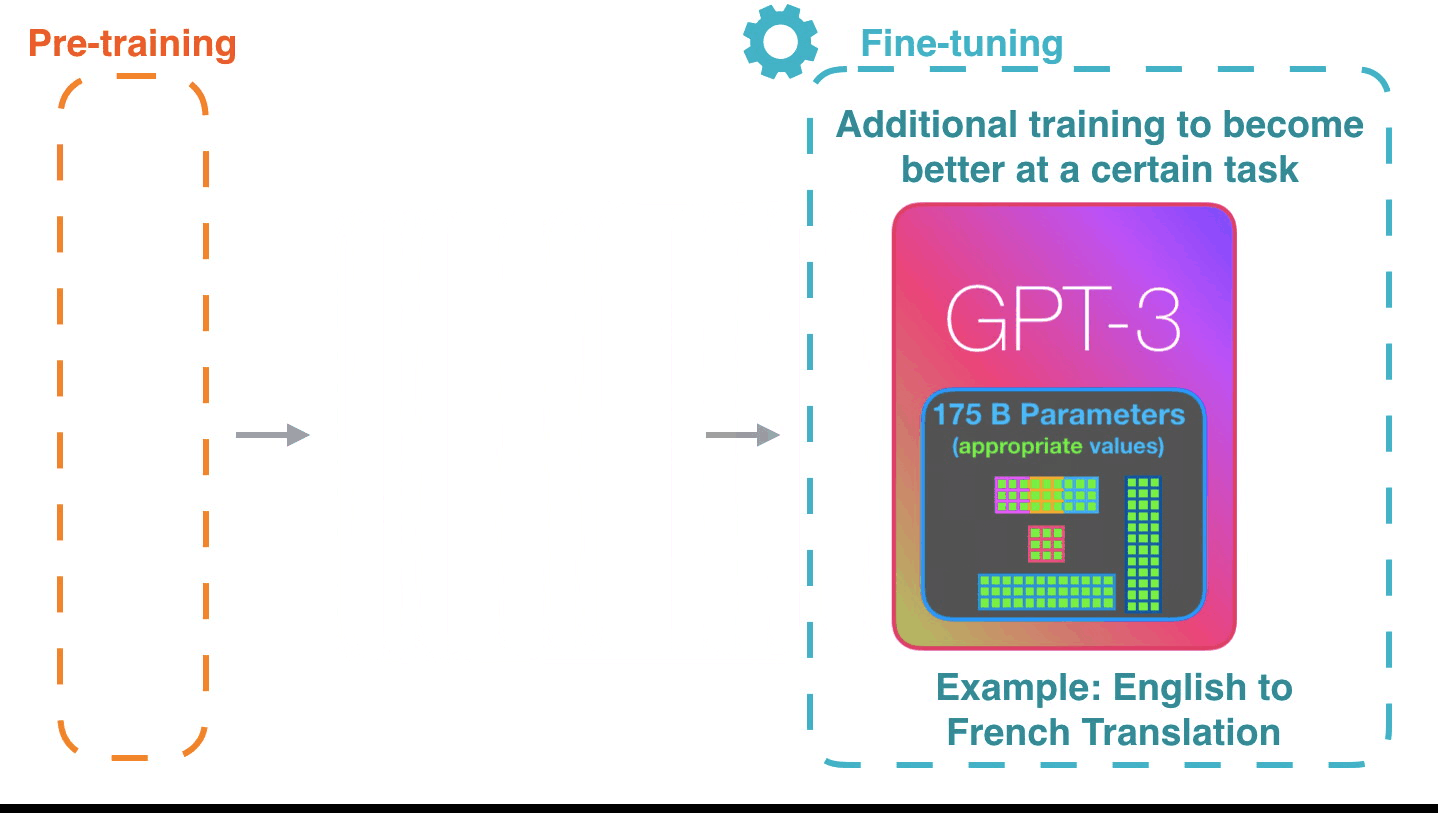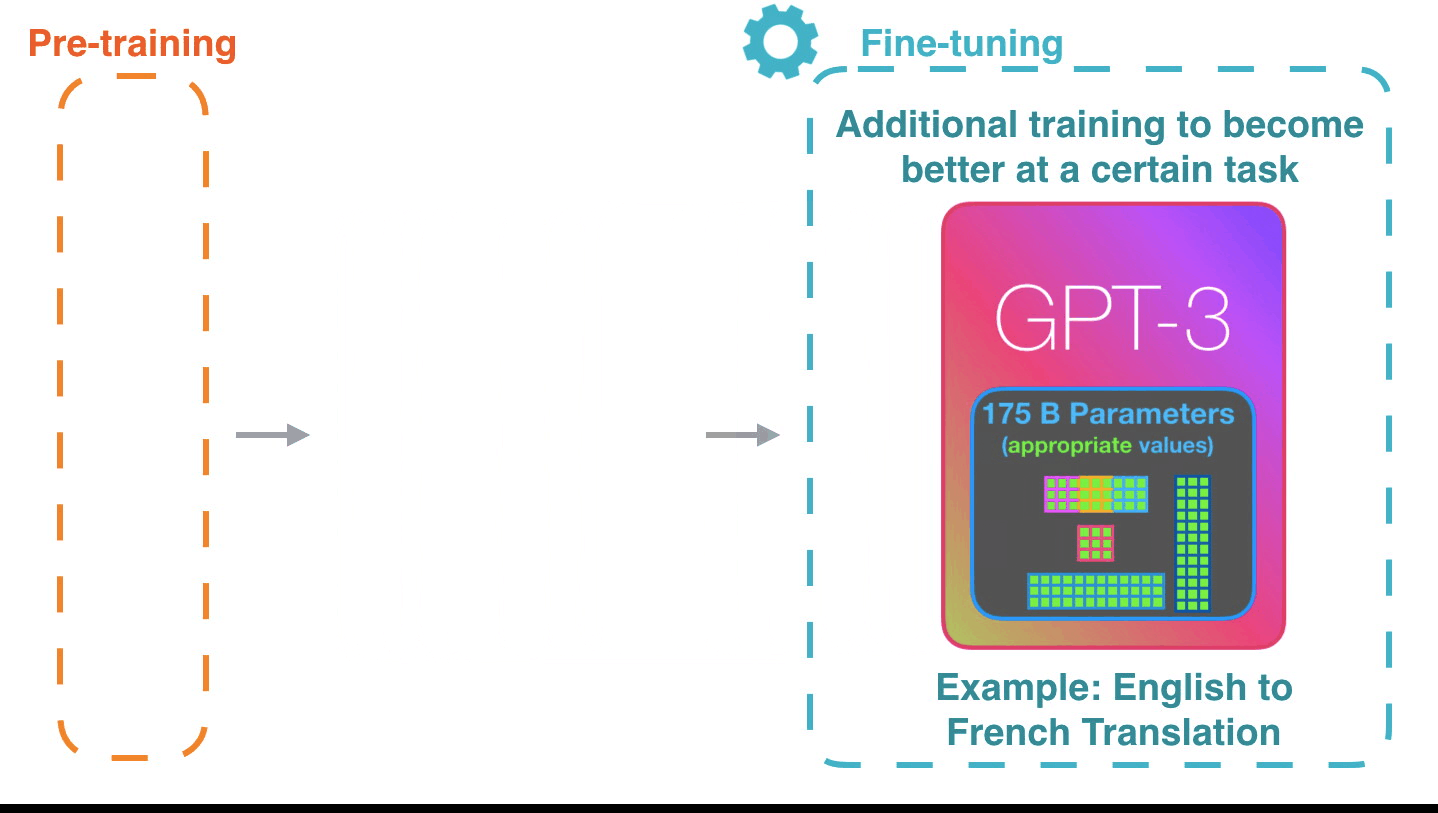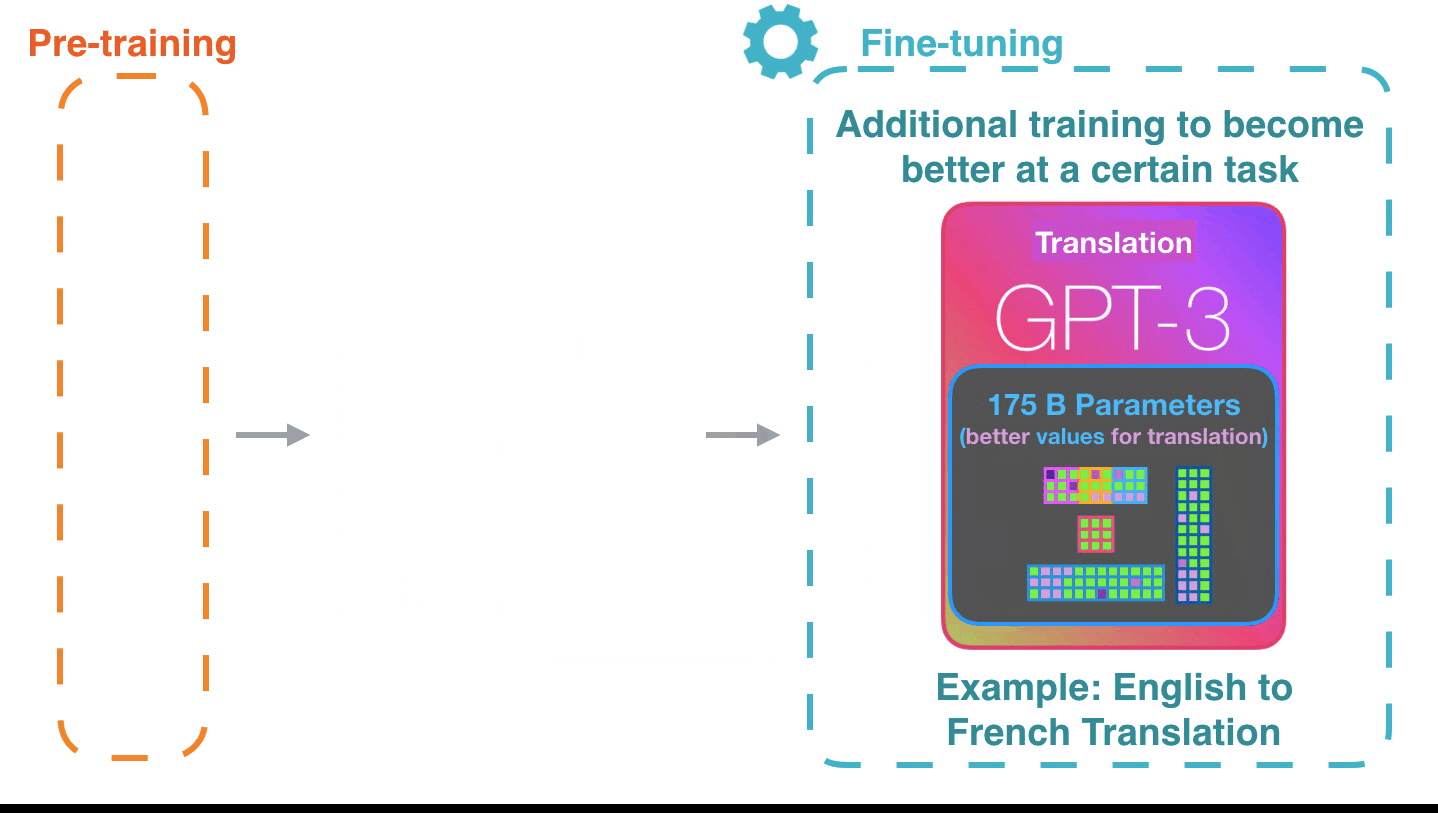
AuroraGPT: General purpose scientific LLM
Broadly trained on a general corpora plus scientific {papers, texts, data}
- Explore pathways towards a “Scientific Assistant” model
- Build with international partners (RIKEN, BSC, others)
- Multilingual English, 日本語, French, German, Spanish
- Multimodal: images, tables, equations, proofs, time series, graphs, fields, sequences, etc
Awesome-LLM
🦙 Issues with “Publicly Available” LLMs
- Trust and Safety:
- Skepticisim about deployment in critical infrastructure
- Correctness and reliability of model outputs
- Transparency:
- Data governance, what was used for pre-training? fine-tuning?
- generally unknown
- What is open source?
- Model weights?
- Pre-training {code, logs, metrics} ?
- Data governance, what was used for pre-training? fine-tuning?
🧪 AuroraGPT: Open Science Foundation Model
📊 AuroraGPT: Outcomes
Datasets and data pipelines for preparing science training data
Software infrastructure and workflows to train, evaluate and deploy LLMs at scale for scientific resarch purposes
- argonne-lcf/Megatron-DeepSpeed
End-to-end training and inference, on any GPU cluster - argonne-lcf/inference-endpoints1
Inference endpoints for LLMs, hosted @ ALCF
- argonne-lcf/Megatron-DeepSpeed
- Evaluation of state-of-the-art LLM Models:
- Determine where they fall short in deep scientific tasks
- Where deep data may have an impact
📚 What do we hope to get?
- Assessment of the approach of augmenting web training data with two forms of data specific to science:
- Full text scientific papers
- Structured scientific datasets (suitably mapped to narrative form)
- Research grade artifacts (models) for scientific community for adaptation for downstream uses1
- Promotion of responsible AI best practices where we can figure them out
- International Collaborations around the long term goal of AGI for science
🌌 Aurora
| Racks | 166 |
| Nodes | 10,624 |
| CPUs | 21,248 |
| GPUs | 63,744 |
| NICs | 84,992 |
| HBM | 8 PB |
| DDR5c | 10 PB |
🤖 ALCF AI Testbed
- ALCF AI Testbed Systems are in production and available for allocations to the research community
- Significant improvement in time-to-solution and energy-efficiency for diverse AI for science applications.
- NAIRR Pilot
Up to ≈ 25× throughput improvement for genomic FMs with 6.5× energy efficiency
👥 Team Leads
🤝 Teams
- Planning
- Data Prep
- Accumulate 20+ T tokens of high-quality scientific text and structured data
- Models / Training1
- Train (entirely from scratch) a series of models on publicly available data
- Evaluation
- Skills, trustworthiness, safety, robustness, privacy, machine ethics
- Post-Training
- Fine-tuning, alignment
- Inference
- Model serving, API development / public-facing web services
- Distribution
- Licensing, generating and distributing artifacts for public consumption
- Communication
📚 Data
✅ Goal: Assemble a large corpus of documents (general and scientific) to train and fine-tune AuroraGPT models
- Challenges: Avoid / detect contamination with benchmarks
- Respect copyright (ACM Digital Library), privacy, and ethical considerations
- Performance Challenges: High throughput data processing
- Converting PDF → text (math formula, figures)
- Convert science information (data) into text (narratives)
- De-duplication (syntactic and semantic) of scientific documents (to avoid memorization, bias)
- Quantity: Considering 20+ Trillion tokens → ≈ 100M papers
- Domains: All (long-term) scientific domains, starting with:
- Material science, Physics, Biology, Computer Science, Climate Science
⏱️ Dataset Processing
- To train a fixed model on trillions of tokens requires:
- Aggregating data from multiple different corpora
(e.g. ArXiv, Reddit, StackExchange, GitHub, Wikipedia, etc.) - Sampling each training batch according to a fixed distribution across corpora
- Building indices that map batches of tokens into these files (indexing)
The original implementation was slow:
- Designed to run serially on a single device
- Major bottleneck when debugging data pipeline at scale
- Aggregating data from multiple different corpora
🚀 Accelerating Dataset Processing: Results
🦜 Model Training
✅ Goals
- Want training runs at scale to be:
- efficient
- stable
- reproducible
- This requires:
- robust data pipelines / file IO
- effectively overlapping compute with communication
- stability across {network, filesystem, machine}
- 3D / Multi-dimensional Parallelism strategies
- Large batch training
- Second order optimizers
- Sub-quadratic attention
- State space models
- Highly optimized GPU kernels
❌ Challenges
- Looong time to train, can be:
- weeks (even months) of continuous training
- order of magnitude longer than typical NN training jobs
- Stability issues:
- failures are expensive (but inevitable)
- stragglers common at scale
- Individual jobs are:
- fragile
- only as good as the worst rank
- one hang or bad worker can crash job
- network / filesystem / other-user(s) dependent
- Cost / benefits of different collective communication algorithms
- depend on optimized / efficient implementations
- Network performance
- Highly optimized GPU kernels
🤔 Evaluating FM Skills for Science
- What to measure?
- Knowledge Extraction, Retrieval, Distillation, Synthesis: LLM is provided a question or instruction and a truthful answer is expected
- Text Grounded: Answers are expected to be fully grounded on peer-reviewed references to support responses
- Reasoning: LLMs are expected to solve deductive (prove a theory or hypothesis from formal logic and observations), inductive (validate / explain observations from theories) problems
- Creativity: A creative answer is expected from a question or instruction
- thoughtful dialogue, coding, etc.
⚖️ Evaluating FM Skills for Science: Criteria
- Criteria for all of the above:
- Correctness of facts
- Accuracy of solutions and inferences
- Reliability consistently good in quality or performance
- Speed how fast to produce a response
- # shots how many examples are needed for good quality
- Extent of prompt engineering
🧬 MProt-DPO: Scaling Results
📓 References
- argonne-lcf /
Megatron-DeepSpeed
For the largest of large language models. - saforem2 /
ezpz
Distributed training, ezpz. 🍋 - 📊 See my other slides at samforeman.me/talks:
❤️ Thank you!
🙏 Acknowledgements
This research used resources of the Argonne Leadership Computing Facility, which is a DOE Office of Science User Facility supported under Contract DE-AC02-06CH11357.
📑 Bibliography
- Refs:
- Wei et al. (2022)
- Animations from The Illustrated Transformer
🎁 Extras
🧬 MProt-DPO: Scaling Results
🚂 Loooooooooong Sequence Lengths
- Working with Microsoft/DeepSpeed team to enable longer sequence lengths (context windows) for LLMs
- See my blog post for additional details
SEQ_LEN for both 25B and 33B models (See: Song et al. (2023))